Abstract
Biological tissues are highly organised and often dynamic. The corresponding spatio- temporal patterns are relevant in many biological and biomedical processes including tissue homeostasis, viral infection or tumour development, and can be studied with imaging techniques, such as light and fluorescence microscopy. Biomedical imaging data provides quantitative information about biological systems, however, mechanisms causing spatial patterning and causalities often remain elusive. It is therefore increasingly appreciated that computational modelling is an essential tool to understand multicellular spatio-temporal processes. Whereas imaging, data analysis and simulation techniques have been in the focus in recent years, one key aspect is just becoming accessible with advanced methods and computational resources: the rigorous parameterisation of multicellular models.
The parameterisation of computational multicellular models from high-throughput and high-content (imaging) data is crucial for the understanding of multicellular processes, the prediction of perturbation experiments and the comparison of competing hypotheses. Substantial progress has been made in recent years by combining rigorous parameter estimation approaches, i.e. Approximate Bayesian Computation (ABC), with high- performance computing methods. However, the available modelling and parameter estimation pipelines for multicellular models are tailor-made to particular applications and difficult – if not impossible – to reuse. There is a growing need for solutions that allow researchers to combine modelling and simulation of multicellular systems with computational tools for data-driven parameter estimation.
The overall aim of FitMultiCell is to build and validate an open platform for modelling, simulation and parameter estimation of multicellular systems, which will be utilised to mechanistically answer biomedical questions based on imaging data.
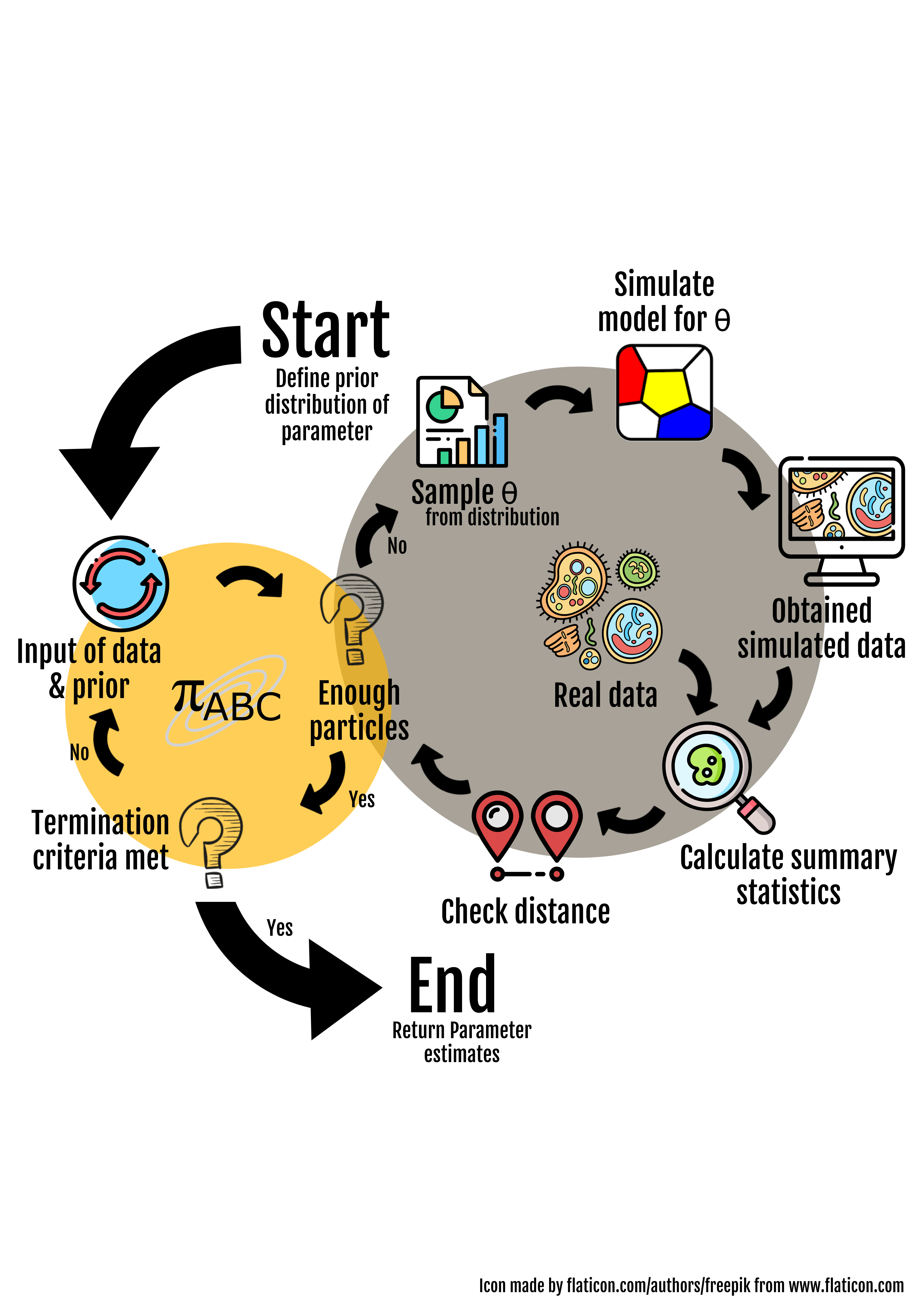
The project Goals
The project aims are the following:
- To develop an integrated computational platform for the data-driven modelling and parameter inference of multicellular systems, based on open-source and extendible software.
- To provide a user-friendly, scalable version of the platform that is easy to use on high performance computing (HPC) platforms including grid and cloud environments.
- To accelerate the simulation of computational multicellular models, which will enable the study of multiscale multicellular processes in 2D and 3D with millions of cells.
- To advance parameter estimation and model selection for multicellular models, which will facilitate a rigorous, data-driven assessment of spatio-temporal processes.
- To validate and apply the platform by studying multicellular models and biomedical imaging data, including novel data on the spread of viral infections and liver disease.
Resources
- Source code: https://gitlab.com/fitmulticell/fit
- Documentation: https://fitmulticell.readthedocs.io/en/latest/
Funding
| FitMultiCell is funded by the German Ministry of Education and Research (BMBF) within the funding program "CompLS – Computational Life Sciences"." |  |